
Research News
Research News
Emergence and mobilization of a novel lincosamide resistance gene lnu(I): From environmental reservoirs to pathogenic bacteria
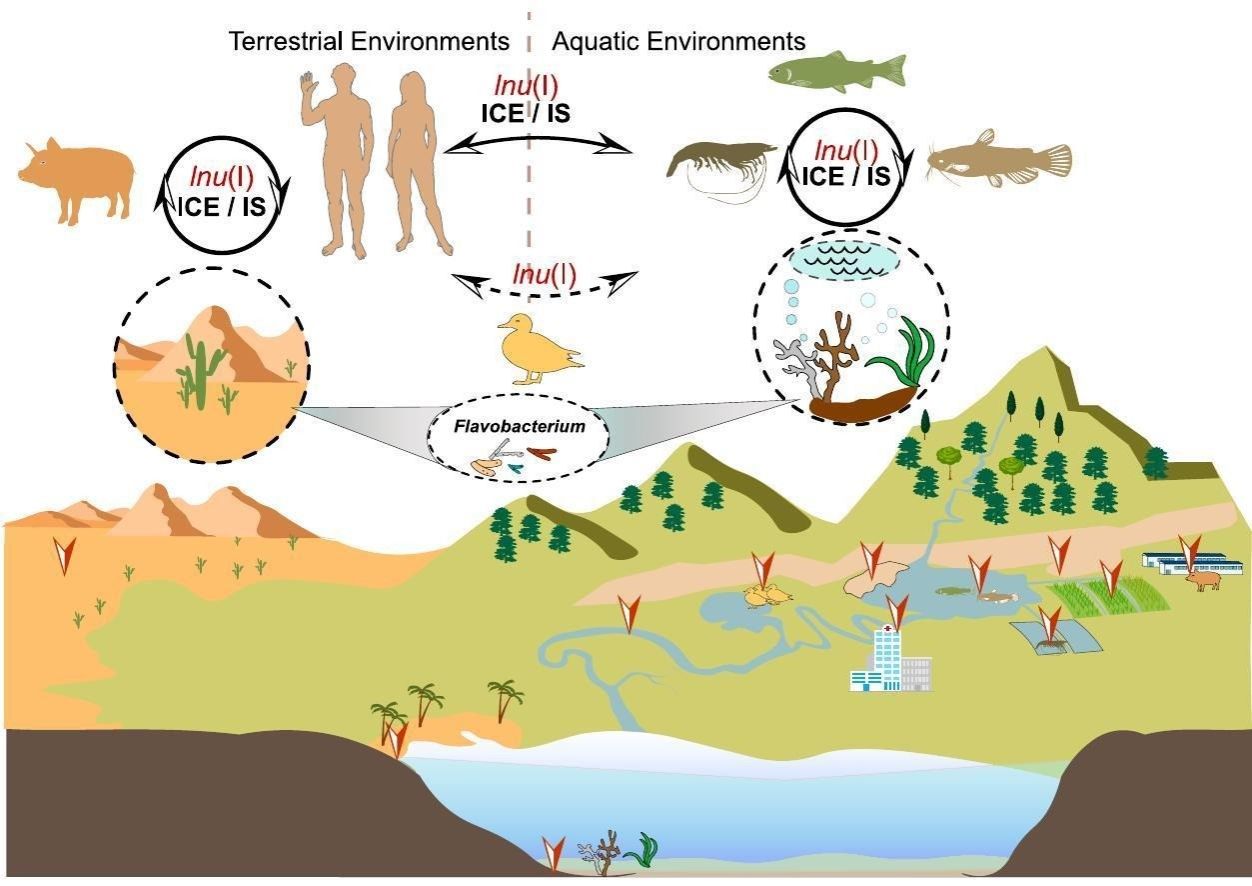
Antimicrobial resistance remains an utmost concern in human and veterinary medicine, impacting humans, animals, and the environment while significantly influencing the principles of One Health. While Riemerella anatipestifer (R. anatipestifer) is recognized as a waterfowl pathogen with multidrug-resistant properties, the specifics of its lincosamide resistance mechanism are inadequately understood. In this study, we identified a novel lincosamide resistance gene, lnu(I), in R. anatipestifer RCAD0121, and investigated its potential origin, transfer mechanisms, and dissemination status through genomic epidemiology. This exhibited 74.80 % amino acid identity with a previously reported gene, lnu(H). PCR analysis revealed lnu(I) prevalence in at least 44 R. anatipestifer isolates collected from multiple provinces in China. Furthermore, genomic mining unveiled 56 lnu(I) sequences within publicly available databases, primarily originating from environmental sources. In addition, members of the family Flavobacteriaceae were the dominant (16/56, 28.57 %) bacteria carrying the lnu(I) gene, with Flavobacterium exhibiting a similar GC content as lnu(I). Notably, specific instances of the lnu(I) gene were linked to mobile genetic elements within human and animal pathogenic bacteria. These findings suggest that Flavobacterium species within the environment could serve as potential ancestral sources of the novel lnu(I) gene, which has undergone mobilization events toward pathogenic bacteria.
https://doi.org/10.1016/j.scitotenv.2023.167400





 028-86296382
028-86296382  No. 211 Huimin Road, Wenjiang District, Chengdu, Sichuan Province
No. 211 Huimin Road, Wenjiang District, Chengdu, Sichuan Province 